To use all functions of this page, please activate cookies in your browser.
my.chemeurope.com
With an accout for my.chemeurope.com you can always see everything at a glance – and you can configure your own website and individual newsletter.
- My watch list
- My saved searches
- My saved topics
- My newsletter
Resampling (statistics)In statistics, resampling is any of a variety of methods for doing one of the following:
Common resampling techniques include bootstrapping, jackknifing and permutation tests. Additional recommended knowledge
BootstrapBootstrapping is a statistical method for estimating the sampling distribution of an estimator by sampling with replacement from the original sample, most often with the purpose of deriving robust estimates of standard errors and confidence intervals of a population parameter like a mean, median, proportion, odds ratio, correlation coefficient or regression coefficient. It may also be used for constructing hypothesis tests. It is often used as a robust alternative to inference based on parametric assumptions when those assumptions are in doubt, or where parametric inference is impossible or requires very complicated formulas for the calculation of standard errors. See also particle filter for the general theory of Sequential Monte Carlo methods, as well as details on some common implementations. JackknifeJackknifing, which is similar to bootstrapping, is used in statistical inferencing to estimate the bias and standard error in a statistic, when a random sample of observations is used to calculate it. The basic idea behind the jackknife estimator lies in systematically recomputing the statistic estimate leaving out one observation at a time from the sample set. From this new set of "observations" for the statistic an estimate for the bias can be calculated and an estimate for the variance of the statistic. Both methods estimate the variability of a statistic from the variability of that statistic between subsamples, rather than from parametric assumptions. The jackknife is a less general technique than the bootstrap, and explores the sample variation differently. However the jackknife is easier to apply to complex sampling schemes, such as multi-stage sampling with varying sampling weights, than the bootstrap. The jackknife and bootstrap may in many situations yield similar results. But when used to estimate the standard error of a statistic, bootstrap gives slightly different results when repeated on the same data, whereas the jackknife gives exactly the same result each time (assuming the subsets to be removed are the same). Richard von Mises was the first to conceive and apply the jackknife, which has some similarity to k-fold and leave-one-out cross-validation techniques. Permutation testsA permutation test (also called a randomization test, re-randomization test, or an exact test) is a type of statistical significance test in which a reference distribution is obtained by calculating all possible values of the test statistic under rearrangements of the labels on the observed data points. In other words, the method by which treatments are allocated to subjects in an experimental design is mirrored in the analysis of that design. If the labels are exchangeable under the null hypothesis, then the resulting tests yield exact significance levels. Confidence intervals can then be derived from the tests. The theory has evolved from the works of R.A. Fisher and E.J.G. Pitman in the 1930s. To illustrate the basic idea of a permutation test,
suppose we have two groups A and B whose sample means
are The test proceeds as follows. First, the difference in means between the two samples is calculated - this is the observed value of the test statistic, T(obs). Then the observations of groups A and B are pooled. From these pooled values, nA observations are sampled at random without replacement. The sample mean for these nA observations is computed, and the sample mean for the remaining nB observations is also computed, and the difference between the resulting sample means is recorded. This process is repeated many times (e.g. 9999 times), until a reliable estimation of the distribution of the test statistic when the null hypothesis is true have been reached. If the only purpose of the test is reject or not reject the null hypothesis, we can as an alternative sort the recorded differences, and then observe if T(obs) is contained within the middle 95% of them. If it does not, we reject the hypothesis of identical probability curves at then 5% significant level. If the number of all possible permutations is small the algorithm can be applied to using all possible ways to sample nA observation from a sample of nA+nB. Relation to parametric testsPermutation tests are a subset of non-parametric statistics. The basic premise is to use only the assumption that it is possible that all of the treatment groups are equivalent, and that every member of them is the same before sampling began (i.e. the slot that they fill is not differentiable from other slots before the slots are filled). From this, one can calculate a statistic and then see to what extent this statistic is special by seeing how likely it would be if the treatment assignments had been jumbled. In contrast to permutation tests, the reference distributions for many popular "classical" statistical tests, such as the t-test, f-test, z-test and chi-squared test, are obtained from theoretical probability distributions. Fisher's exact test is a commonly used test for evaluating the association between two dichotomous variables, that is a permutation test. When sample sizes are large, the Pearson's chi-square test will give accurate results, but for small samples the chi-square reference distribution can't be assumed to give a correct description of the probability distribution of the test statistic, and in this situation the use of Fisher’s exact test becomes more appropriate. A rule of thumb is that the expected count in each cell of the table should be greater than 5 before Pearson's chi-squared test is used. Permutation tests exist in many situations where parametric tests do not. For example, when deriving an optimal test when losses are proportional to the size of an error rather than its square. All simple and many relatively complex parametric tests have a corresponding permutation test version that is defined by using the same test statistic as the parametric test, but obtains the p-value from the sample-specific permutation distribution of that statistic, rather than from the theoretical distribution derived from the parametric assumption. For example, it is possible in this manner to construct a permutation t-test, a permutation chi-squared test of association, a permutation version of Aly's test for comparing variances and so on. The major down-side to permutation tests are that
ExamplesAdvantagesPermutation tests exist for any test statistic, regardless of whether or not its distribution is known. Thus one is always free to choose the statistic which best discriminates between hypothesis and alternative and which minimizes losses. Permutation tests can be used for analyzing unbalanced designs (http://tbf.coe.wayne.edu/jmasm/vol1_no2.pdf) and for combining dependent tests on mixtures of categorical, ordinal, and metric data (Pesarin, 2001). Before the 1980s, the burden of creating the reference distribution was overwhelming except for data sets with small sample sizes. But since the 1980s, the confluence of cheap fast computers and the development of new sophisticated path algorithms applicable in special situations, made the application of permutation test methods practical for a wide range of problems, and initiated the addition of exact-test options in the main statistical software packages and the appearance of specialized software for performing a wide range of uni- and multi-variable exact tests and computing test-based "exact" confidence intervals. LimitationsAn important assumption behind a permutation test is that the observations are exchangeable under the null hypothesis. An important consequence of this assumption is that tests of difference in location (like a permutation t-test) require equal variance. In this respect, the permutation t-test shares the same weakness as the classical Student’s t-test. A third alternative in this situation is to use a bootstrap-based test. Good (2000) explains the difference between permutation tests and bootstrap tests the following way: "Permutations test hypotheses concerning distributions; bootstraps tests hypotheses concerning parameters. As a result, the bootstrap entails less-stringent assumptions." Of course, bootstrap tests are not exact. Monte Carlo testingAn asymptotically equivalent permutation test can be created when there are too many possible orderings of the data to conveniently allow complete enumeration. This is done by generating the reference distribution by Monte Carlo sampling, which takes a small (relative to the total number of permutations) random sample of the possible replicates. The necessary size of the Monte Carlo sample depends on the need for accuracy of the test. If one merely wants to know if the p-value is significant, sometimes few as 400 rearrangements is sufficient to generate a reliable answer. However, for most scientific applications the required size is much higher. For observed p=0.05, the accuracy from 10,000 random permutations is 0.0056 and for 50,000 it is 0.0025. For observed p=0.10, the corresponding accuracy is 0.0077 and 0.0035. Accuracy is defined from the binomial 99% confidence interval: p +/- accuracy See also
BibliographyIntroductory statistics
Resampling methods
Bootstrapping
Permutation testOriginal references:
Modern references:
Computational methods:
References
Software
|
|
| This article is licensed under the GNU Free Documentation License. It uses material from the Wikipedia article "Resampling_(statistics)". A list of authors is available in Wikipedia. |





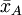 and
and 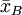 ,
and that we want to test, at 5% significance level, whether they come from the same distribution.
Let
,
and that we want to test, at 5% significance level, whether they come from the same distribution.
Let 

