Good vibrations help reveal molecular details
Five years of hard work and a little "cosmic luck" led Rice University researchers to a new method to obtain structural details on molecules in biomembranes.
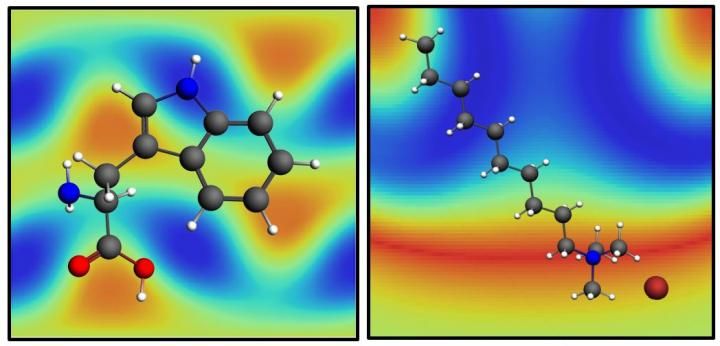
The molecules tryptophan, left, and decyltrimethylammonium bromide, right, over their SABERS maps. SABERS, a new analysis method developed at Rice University, is able to obtain structural details of molecules in lipid membranes near gold nanoparticles without molecular tags.
Hafner Lab/Rice University
The method by the Rice lab of physicist Jason Hafner combines experimental and computational techniques and relies on the plasmonic properties of gold nanoparticles. It takes advantage of the nanoparticles' unique ability to focus light on very small targets.
The researchers call their protocol SABERS, for structural analysis by enhanced Raman scattering, and say it could help scientists who study amyloid interactions implicated in neurodegenerative disease, the neuroprotective actions of fatty acids and the function of chemotherapy agents.
Their method extracts the location of specific chemical groups within the molecules by locating their characteristic vibrations. When a laser activates plasmons in the nanoparticles, it amplifies vibrationally scattered light from nearby molecules, a phenomenon called surface-enhanced Raman scattering (SERS). The enhancement is sensitive to exactly where the molecule sits relative to the nanoparticle.
"Molecules can vibrate in many different ways, so we have to assign a 'center of vibration' to each one," Hafner said. "If you watch some part of a molecule vibrating, you can visualize where it occurs, but we also had to find a mathematical way to describe it."
SERS spectra are notoriously difficult to untangle, so the full SABERS method also requires unenhanced spectral measurements and theoretical calculations of both the nanorod optics and the molecular properties, he said.
Hafner and his team tested their technique on three structures: surfactant molecules that come with gold nanorods, lipid molecules that form membranes on gold nanorods and tryptophan, an amino acid that settles into the membrane.
"We found that the surfactant layer is tilted by 25 degrees, which is interesting because it explains why other measurements found that the layer appears thinner than expected," Hafner said.
Lipids easily replace surfactants on nanorods since they end in the same chemical structure. By comparing vibrations of that structure in the lipid headgroup to a double bond in the tail, SABERS found the correct orientation and thickness of the lipid bilayer membrane. "It's just cosmic luck that a lipid ends in a perfectly symmetric structure that vibrates and is Raman active and loves to sit on a nanorod," Hafner said.
The researchers also used SABERS to locate tryptophan in the lipid bilayer. "It's very bright, spectroscopically, and easy to see," he said. "In real biological structures, tryptophan is just a small residue attached to a much larger protein. However, tryptophan helps anchor the protein to the membrane, so researchers want to know where it prefers to sit."
Next, Hafner wants to analyze bigger molecules. "In principle, through spectroscopic tricks, we could take this to larger structures, and perhaps even find every residue in a protein to get the whole structure. That's futuristic, but it's where we think we can go with it," he said.
Original publication
Other news from the department science

Get the chemical industry in your inbox
From now on, don't miss a thing: Our newsletter for the chemical industry, analytics, lab technology and process engineering brings you up to date every Tuesday and Thursday. The latest industry news, product highlights and innovations - compact and easy to understand in your inbox. Researched by us so you don't have to.