New PET Hydrolase
Enzyme in salivary microbes decomposes PET-based plastics
Advertisement
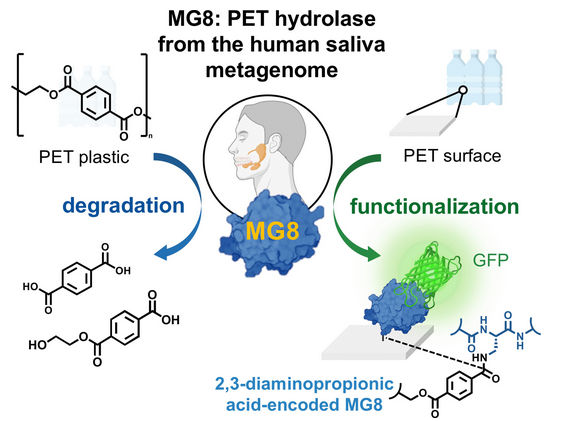
Human saliva may contain an enzyme which can decompose the plastic polyethylene terephthalate (PET). Researchers found the promising enzyme, a hydrolase, in a database containing human metagenome samples. As they report in the journal Angewandte Chemie, this newly discovered hydrolase performs better than many other known bacterial PET hydrolases. It can be produced using biotechnological methods and could be put to use in plastic recycling or for functionalizing plastics, the authors add.
© Wiley-VCH
Landfill sites and harbors are known to be particularly promising sites for finding bacteria that have adapted to consume or make use of plastics. These bacteria have evolved enzymes, known as PET hydrolases, which can break PET down into smaller molecules. Chayasith Uttamapinant from the Vidyasirimedhi Institute of Science and Technology (VISTEC) in Rayong, Thailand, and Worawan Bhanthumnavin from Chulalongkorn University, Bangkok, Thailand, and colleagues, have now discovered the first enzyme to decompose PET from a rather more surprising source: the genome of microbial communities in human saliva.
The researchers believe that, because humans consume vast amounts of food which has been packaged using PET, microbes in the saliva, or the GI tract, may have evolved to digest microplastics. The team uncovered the new hydrolase, which they named MG8, while searching a public metagenome database containing samples from seawater and human saliva, and were able to attribute the likely source of the enzyme to Gram-negative bacteria that may reside in human saliva. These bacteria are similar to strains found near the “Pacific trash vortex”, which have also evolved to produce PET hydrolases.
They first needed enough material to perform their experiments, and so they modified a bacterium that can be cultured in laboratories to produce the enzyme. They easily recovered an active form of the enzyme, able to decompose PET, from a denatured form that can be isolated in large amounts. The researchers highlight that this shows great promise for scaling up in the future.
Aside from the prospects for recycling scalability, the team also foresee another use of MG8. They discovered that not only can it decompose PET with ease but, with a small modification, it can also bind to it highly effectively. To achieve this, they modified the protein sequence by replacing one of the naturally occurring amino acids (serine) at the active site with an unnatural amino acid, DAP. The modified enzyme immediately adhered to PET powder. This could be used as a vehicle for functionalizing PET surfaces, increasing the versatility of PET in medical devices, for example, and enhancing the versatility of recycled PET.
Despite the promise of MG8 in plastic recycling and functionalization, the team acknowledge that MG8, like other PET hydrolases, still needs some work. For the time being, consumer-grade PET plastics with high crystallinity cannot be decomposed using this hydrolase. Therefore, further research will be necessary to reach the stage where a whole plastic water bottle can be dissolved in a simple solution containing the enzyme.